Life Sciences – Group of Thomas Sokolowski
Research focus
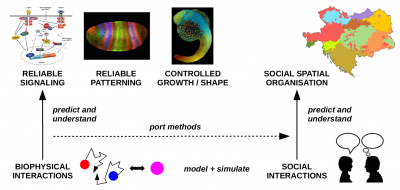
We ask: How can cells reliably generate, sense, transmit and process biochemical signals using limited molecular resources implying a high degree of noise? How can tissues and organisms develop accurately and reproducibly based on such noisy biochemical processes? What are the biophysical mechanisms that evolved to grant cells and tissues the ability to control molecular noise for reliable information processing? How do spatial effects and processes, such as molecular diffusion and active transport on the cytoskeleton, impact on these mechanisms? And, finally, can we port our mechanistic understanding of noise control inside organisms to elucidate spatial-stochastic processes on the multi-organism / population scale?
We pursue our research at the interface of computational and statistical physics, quantitative biology, biochemistry, information theory, and mathematical optimization. Our systems of interest cover multiple spatial scales ranging from few biochemical particles in single cells up to tissues consisting of thousands of cells and, as an emerging theme, organism populations.
In the following, we briefly portray our current main research themes.
Interessiert?
Students and young researchers interested in physics-driven computational approaches for studying the robustness of biological systems are warmly welcome to contact us for potential projects or internships.
Please also check our teaching activities at Goethe University for interesting courses and seminars in biophysics, computational physics and scientific programming.
Current Research Projects
GEWEBEENTWICKLUNG IN FLIEGEN- UND MAUSEMBRYOS
TISSUE DEVELOPMENT IN FLY AND MOUSE EMBRYOS
Perhaps one of the most intriguing and astonishing processes in biology is the development of an embryo from its genetic blueprint initially contained in a single fertilized cell. While the basic principles of embryo development today are common knowledge, it is often overseen that—microscopically—embryogenesis is based on biochemical processes that operate at low molecular numbers, and therefore are intrinsically stochastic. This implies a high degree of noise in the protein signals driving the early differentiation of developing cells, contrasting with the outstandingly high precision and reproducibility observed in many embryo systems. Moreover, this precision is attained in remarkably short developmental time, excluding temporal averaging as a noise control mechanism. How can early embryogenesis be so reliable and fast in spite of limited molecular resources?
Our research aims at uncovering biophysical and biochemical mechanisms that suppress noise in early tissue development in order to make it reliable and robust. To this end we develop biophysically realistic spatial-stochastic models of emerging embryos and augment their computational efficiency by accurate coarse-graining and smart simulation algorithms. This allows us to explore large sets of biophysical parameters in order to find model solutions that are optimal in suppressing biological noise.
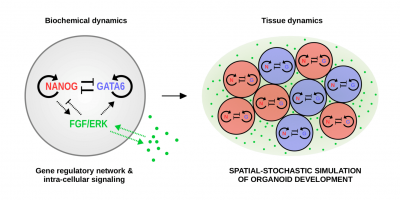
We currently focus on two particular systems: the gap gene system in the early embryo of the fruit fly Drosophila melanogaster, and the early, preimplantation-stage mouse embryo. While these two systems both contain regulatory networks with features that appear commonplace in development, they fundamentally differ in the order of developmental events: While in the early fly first, a (transient) tissue structure is established and subsequently, different cell fates are acquired simultaneously by reading out positional information provided by morphogens, the preimplantation mouse embryo is highly dynamic and early cell fates emerge as the tissue is built up by cell divisions and reorganizations. By comparing noise control mechanisms between the two systems, we aim at disentangling recurrent noise-control strategies in development from those that appear organism-specific and understanding the circumstances under which particular mechanisms are beneficial.
SPATIAL EFFECTS IN TRANSCRIPTION FACTOR BINDING
Unarguably, transcription factor binding resulting in activation or suppression of gene expression forms the core basis of gene regulation. While in the past decades numerous studies have uncovered the principles of gene regulation by transcription factors, its fundamentally stochastic nature, and many mechanisms that evolved to cope with it, our knowledge of how transcription regulation can happen reliably under limited molecular resources remains incomplete. This holds in particular for the eukaryotic realm, where target regions regulated by transcription factor oftentimes show more complex structures than one would deem necessary, including numerous oftentimes identical binding sites and distant enhancers (in addition to promoter regions) that, seemingly unnecessarily, waste molecular resources. Moreover, recent studies hint at a novel regulatory paradigm in eukaryotic systems in which the classical "search and bind" mechanism of transcription factors seems to step back in favor of highly dynamical "transcription factor hubs", i.e. "clouds" of transcription factors that accumulate in the relevant, regulated regions of the DNA by transient binding at a fast turnover. It remains unclear why such mechanisms of transcriptional regulation would be beneficial and superior to the classical "search and bind" mechanism.
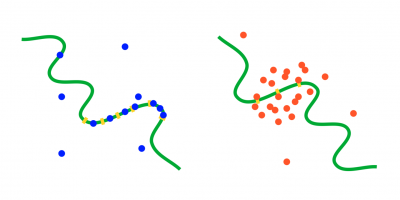
We ask how particular binding site distributions on the promoter regions affect binding affinity, and how a defined, persistent regulatory state can be created by dynamical clouds of transcription factors in the first place. We also aim at understanding the advantages and disadvantages of these binding mechanisms as compared to the classical "search and bind" process. To this end, we combine particle-based simulations (mainly using eGFRD2, an exact and efficient event-driven particle-based simulator for reaction-diffusion problems) and mathematical calculations.
SPATIAL-STOCHASTIC PROCESSES IN POPULATIONS
The past decades have seen ample progress both in modeling / simulating and understanding spatial-stochastic effects in cellular systems. However, spatial-stochastic effects, in particular diffusion, also play a key role in spatial-stochastic processes on the scale of populations and even societies. Prominent examples are the spread of languages or social traits and epidemics. By relying on stochastic mechanisms of generation/degradation and transport of the relevant traits, these systems are remarkably similar to spatial biochemical systems. Based on our experience in biochemical modeling, we, therefore, explore whether and how our methods and insights in cellular biology can be ported to the population scale and thereby aid our understanding of the corresponding stochastic processes and the mechanisms that can control them. Our favorized candidate processes are spatial language and epidemics spread, in which diffusive motion and random interactions play a key role, akin to biochemical systems.